
I’m an advocator for learning Python, especially as a data practitioner. Here, you can read my prior post on why Python should be your first programming language.
If you’re in a data-driven company, then you understand the importance of having quality data to make decisions that impact the direction and bottom line of the company. However, if the data is bad, so are the interpretations. It’s the classic case of garbage in, garbage out.
There are multiple points where data can be corrupted and/or fixed. For example, the initial data collection from IoT devices can be corrupted at any moment. Or the pipeline that a data engineer creates can enhance the quality of the data coming in.
This blog is written as an awareness for those who use exploratory data analysis (EDA) more often in their work. Even though Python is a versatile language that’s usually easier to pick up, there are pitfalls that you should be aware of.
You may wonder where and how data can be corrupted once it is ready for an analyst. It’s easier than you think.
Here’s a hint: mutation.
And no, I’m not talking about Teenage Mutant Ninja Turtles or Leela from Futurama 😉.
What is a DataFrame?
As a data analyst or data scientist, you’re probably familiar with either Pandas or PySpark. Both tools have a data structure called DataFrame, which is similar to a table in excel.
There are rows and columns, where the rows indicate individual records, and the columns are the attributes of the data.
The cool thing is that DataFrames are immutable. Immutable objects are objects that cannot be changed after being created. An example of an immutable object is a string. You can’t change/update a string once it’s been created; you can, however, reassign its associated variable to a new string object.
Take note of that last sentence.
⚠️ Pitfall #1: Reassignment
Python allows you to reassign a variable to another object easily. Variables are not bound to a type. So a variable named age assigned to the int 38 can later be set to the string "38", and you wouldn’t get an assignment error.
age = 38
print(type(age))
#----- output: <class 'int'> -----#
age = "38"
print(type(age))
#----- output: <class 'str'> -----#If you’re working with DataFrames, it can be easy to reassign a new DataFrame to an existing variable. It’s not necessarily a bad thing. Sometimes I do this when cleaning my data before beginning EDA.
from pyspark.context import SparkContext
from pyspark.sql.session import SparkSession
import pyspark.sql.functions as F
sc = SparkContext.getOrCreate()
spark = SparkSession(sc)
data = spark.read.parquet("/path-to-file/file.parquet")
data = data.dropDuplicates(["user-id", "date"])
data = data.withColumn("mph", F.col("meters_per_sec") * 2.237)
data = data.filter(data.mph >= 200)In the toy example, the three following functions I called (dropDuplicates, withColumn, and filter) all return a new DataFrame, and I’m reassigning data to each new DataFrame object. There’s a total of 4 assignments for the variable data.
I can get unimaginative or lazy, so I’ll reuse a variable name to make life easier for me. But this could lead to weird insights.
On line 10 of the code snippet, I used a filter. My analysis would be warped if I forgot that the filter was applied or mistakenly reassigned the data variable to the new filtered DataFrame object.
I can’t have good insights on records where the mph is less than 200 since that data was initially excluded!
How to Prevent Pitfall #1
The reassignment pitfall can be prevented by using descriptive variable names. Using descriptive variable names is a good coding practice in general; this will help you and your peers when they go over your code.
raw_data = spark.read.parquet("/path-to-file/file.parquet")
cleaned_data = raw_data.withColumn("mph", F.col("meters_per_sec") * 2.237) \
.dropDuplicates(["user-id", "date"])
mph_gt_200 = cleaned_data.filter(data.mph >= 200)In this new code snippet, I’ve created three new variables and combined the withColumn and dropDuplicates functions.
⚠️ Pitfall #2: Slicing
This pitfall is associated with Pandas in particular.
With Pandas, there are a few ways to slice your data and return a subset. Most people use either .loc() or [] when slicing their data.
courses = {'Courses':['Calc I', 'Diff EQs', 'Intro to Java', 'Stats 101'],
'NumberOfStudents':[18, 15, 35, 20],
'Duration':['15 Weeks','15 Weeks','15 Weeks','15 Weeks']}
df = pd.DataFrame.from_dict(courses)
df2 = df.loc[:1]
df2.NumberOfStudents[0] = 180Pop Quiz: will changing the number of students enrolled in Calc I for df2 impact the original DataFrame, df?
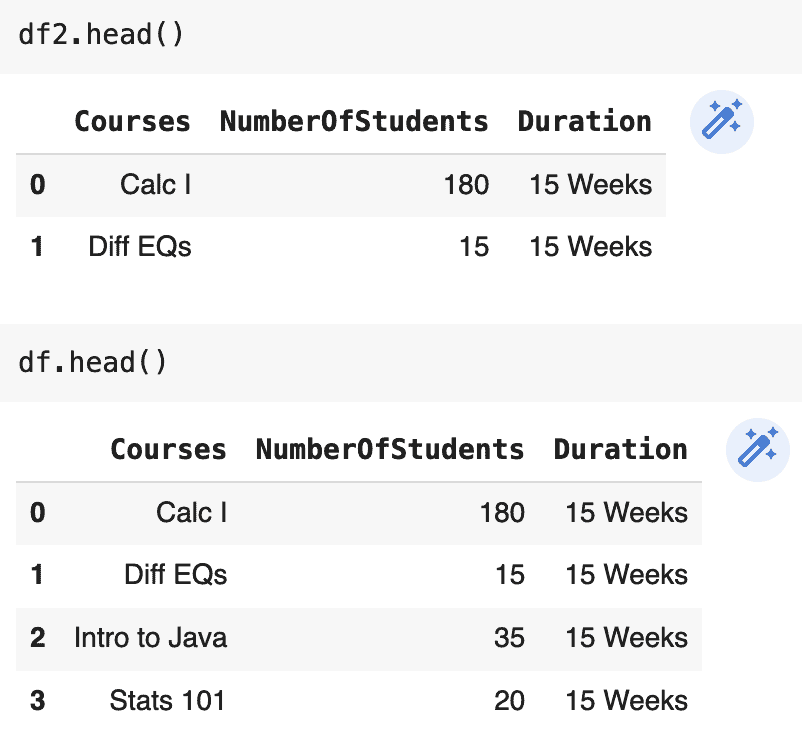
The answer is yes. Changing df2 does impact df; this is because the behavior of indexing and slicing is unpredictable. Sometimes a copy is returned, while a view is returned at other times. For example:
courses = {'Courses':['Calc I', 'Diff EQs', 'Intro to Java', 'Stats 101'],
'NumberOfStudents':[18, 15, 35, 20],
'Duration':['15 Weeks','15 Weeks','15 Weeks','15 Weeks']}
df = pd.DataFrame.from_dict(courses)
df2 = df.loc[:, df.columns]
df2.NumberOfStudents[0] = 180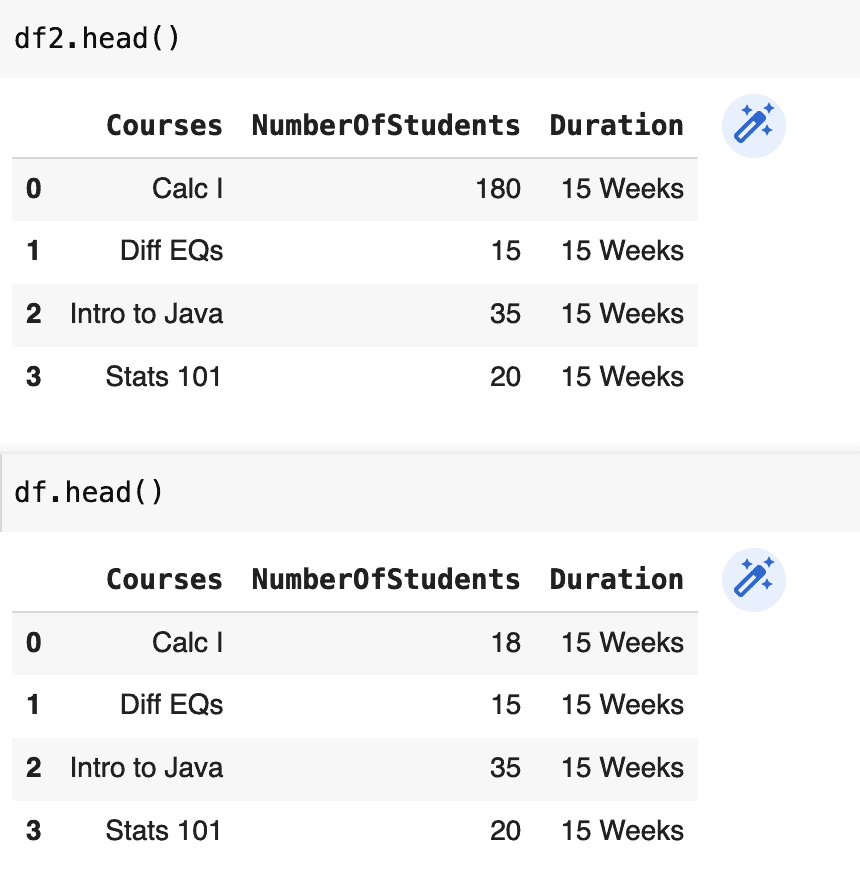
The prior example was a view and thus affected the original DataFrame, while the latter example was a copy and did not affect the original DataFrame.
How to Prevent Pitfall #2
Remember this motto: when in doubt, copy out! If you aren’t sure whether or not your slicing and indexing results in a view or a copy, err on the side of caution by explicitly adding the copy function to ensure you aren’t altering the original DataFrame.
courses = {'Courses':['Calc I', 'Diff EQs', 'Intro to Java', 'Stats 101'],
'NumberOfStudents':[18, 15, 35, 20],
'Duration':['15 Weeks','15 Weeks','15 Weeks','15 Weeks']}
df = pd.DataFrame.from_dict(courses)
df3 = df[df.NumberOfStudents > 18].copy()
df3.NumberOfStudents = [180, 30]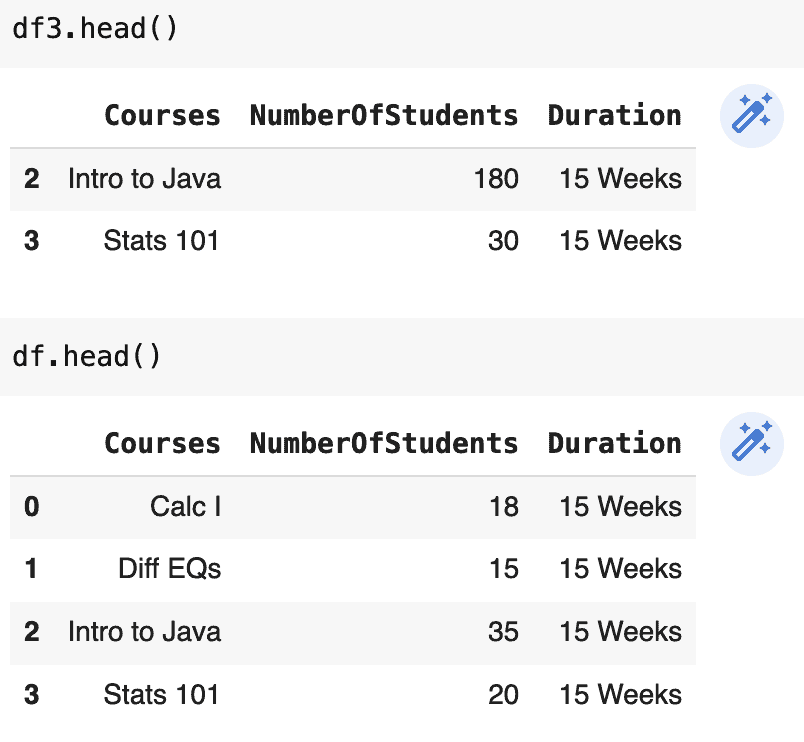
Slicing and indexing are some of the most common operations to perform when doing EDA work. Unfortunately, data could be corrupted without realizing it if we alter our slices and don’t realize the changes are propagated to the original DataFrame.
This type of mutation is a silent killer. Some call it a logic error since the code will run fine, but our calculations and analyses will be wrong!
⚠️ Pitfall #3: Setting inPlace=True
Pandas allow you to alter a DataFrame without returning a new DataFrame. Several functions have a parameter called inPlace, which is usually set to False by default, but you can change it to True.
Pitfall 3 is like the opposite of pitfall 2 because when you set inPlace=True, you are deliberately mutating your DataFrame, and previous slices derived from the original DataFrame will also be altered.
Changing a DataFrame in place is something to really consider because the consequences can lead to unnecessary work and headaches when trying to resolve future bugs.
Perhaps you’re like me and don’t like creating multiple variable names to store intermediate DataFrames. Similar to pitfall 1, I’d still say you should create descriptive variable names!
I usually only set inplace=True when I need to reset the index of a DataFrame after performing an operation like dropping duplicates, using a boolean mask, or using .query(). I feel weird working with a DataFrame that has indices which are out of order 🤷🏾♀️.
How to Prevent Pitfall #3
Honestly, if you’re new to manipulating DataFrames stay away from setting inPlace to True. Once you’re comfortable with what you’re doing, you can use that keyword as you see fit. However, the creators of Pandas are looking to deprecate that parameter, and they discourage us from using it. Ooops, I’m guilty 🤭.
All the pitfalls I’ve listed above are obstacles that I’ve encountered. I wrote this article for you to be aware that data corruption can happen through the functions we call all the time.
Do better than me 😭! But if you’ve made any of the above mistakes, that’s okay too. As long as you’ve learned from that experience, you’ll be less likely to make the same error, and you’re improving your coding/EDA skills ✨!
